|
DICE: Discovering Informative Co-occurring
Elements
 Biological networks connect related genes together
based on their physical interactions, functional relationships,
co-expression, genetic interactions, etc. Many gene features tend to co-occur
in a biological network. For example, proteins that are connected to each
other in a network of physical interactions usually share similar functions.
Similarly, genes that are co-expressed (and thus are connected in a
co-expression network) often possess similar cis-regulatory elements in their upstream and/or downstream
regions. Also, since domain-peptide interaction is a major mediator of
protein-protein interactions, in a protein-protein interaction network
certain domains may often be accompanied by particular peptides, thus forming
a “pair of co-occurring features” within that network. DICE is a universal
approach for identification of such pairs of co-occurring features in
biological networks across all data types. The general framework that DICE applies
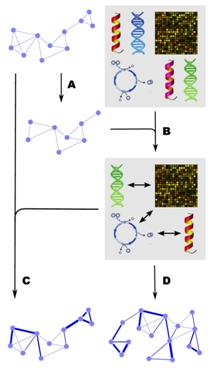
for finding co-occurring pairs of features is depicted in the right picture.
Given a biological network, co-occurring features can be found from a wide
range of data types. Paralogs are first removed from the network (A), and
then different gene features are examined for co-occurrence in the network
(B), resulting in a set of co-occurring features. This set may include
protein domains, GO terms, expression profiles, protein phylogeny
information, codon usage values, nucleic acid and protein motifs that are
discovered de novo based on
co-occurrence in the network, etc. These co-occurring features can be used to
score the interactions (C) and subsequently improve the quality of the
network. Alternatively, they can be used for prediction of novel interactions
(D). Biological networks connect related genes together
based on their physical interactions, functional relationships,
co-expression, genetic interactions, etc. Many gene features tend to co-occur
in a biological network. For example, proteins that are connected to each
other in a network of physical interactions usually share similar functions.
Similarly, genes that are co-expressed (and thus are connected in a
co-expression network) often possess similar cis-regulatory elements in their upstream and/or downstream
regions. Also, since domain-peptide interaction is a major mediator of
protein-protein interactions, in a protein-protein interaction network
certain domains may often be accompanied by particular peptides, thus forming
a “pair of co-occurring features” within that network. DICE is a universal
approach for identification of such pairs of co-occurring features in
biological networks across all data types. The general framework that DICE applies
for finding co-occurring pairs of features is depicted in the right picture.
Given a biological network, co-occurring features can be found from a wide
range of data types. Paralogs are first removed from the network (A), and
then different gene features are examined for co-occurrence in the network
(B), resulting in a set of co-occurring features. This set may include
protein domains, GO terms, expression profiles, protein phylogeny
information, codon usage values, nucleic acid and protein motifs that are
discovered de novo based on
co-occurrence in the network, etc. These co-occurring features can be used to
score the interactions (C) and subsequently improve the quality of the
network. Alternatively, they can be used for prediction of novel interactions
(D).
DICE
Developers:
Hamed Shateri Najafabadi
Yuan Mao
Reza Salavati
Last updated on 3/23/2010 12:02:31 PM
|