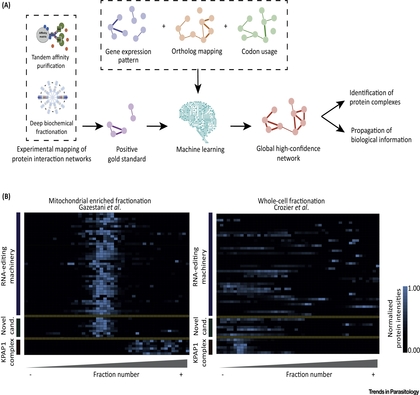
Systems analysis of trypanosomatid genomes
Our lab is focused on the development of novel computational and experimental methods for the functional and structural characterization of trypanosomatid genomes. We use systems biology of trypanosomatid pathogens to identify key genes / pathways and determine their function.
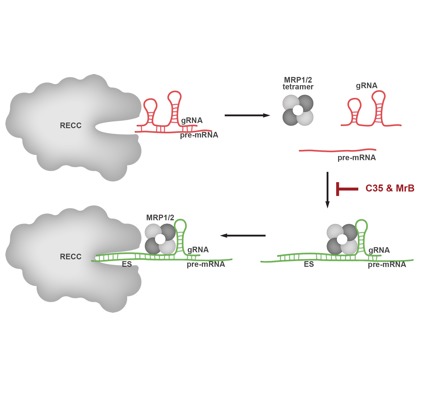
Dynamics and developmental regulation of the editosome function in trypanosomatids
Another major research topic in our lab is the functional characterization of key trypanosomatid proteins that are involved in post-transcriptional gene regulation.
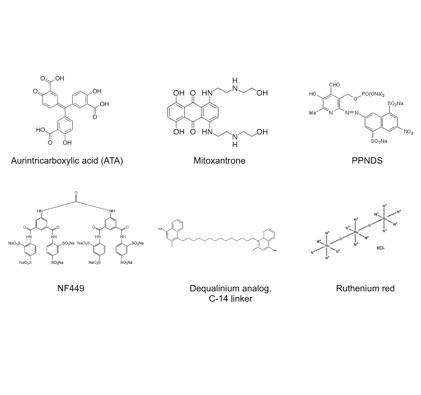
Targeting the essential pathways in trypanosomatids for drug discovery
We are involved in the development of high-throughput methods for the identification of novel chemical inhibitors of essential trypanosomatid proteins. We have identified potential inhibitors for RNA editing that kill T. brucei in vitro with a proposed mechanism of inhibition.